The $1,000 Challenge!
To go straight to the actual challenge click here.
Crack the code! The authors announce the HEGP (heegeepee;
hiːdʒiːpiː) Challenge,  with a $1,000 (one thousand dollar) prize for
the individual or group who can crack HEGP encrypted data. HEGP, when
proven solid, will have a large impact on the way human genetics is
pursued today because it will allow for sharing genotype data while
preserving privacy of the individual. Not only is sharing data
required for reproducible Science, there is also a large interest in
hosting data on laptops and servers that need not be HIPAA compliant.
with a $1,000 (one thousand dollar) prize for
the individual or group who can crack HEGP encrypted data. HEGP, when
proven solid, will have a large impact on the way human genetics is
pursued today because it will allow for sharing genotype data while
preserving privacy of the individual. Not only is sharing data
required for reproducible Science, there is also a large interest in
hosting data on laptops and servers that need not be HIPAA compliant.
On this page we chat about cracking HEGP, Rubik cube, DNA, strawberries and Enigma.
To go straight to the actual challenge click here.
Relevance
With a publication in Genetics we present a novel Homomorphic encryption method named HEGP that allows for sharing of genotypes and phenotypes in the context of Genome Wide Association (GWA) studies. Importantly sharing but without giving away private information and making individuals identifiable. For more information check out a privacy researcher's perspective and a recent follow up paper on Using encrypted genotypes and phenotypes for collaborative genomic analyses to maintain data confidentiality by Tianjing Zhao, Fangyi Wang, Richard Mott, Jack Dekkers, and Hao Cheng, Genetics. 2024 Mar 6;226(3):iyad210. doi: 10.1093/genetics/iyad210. PMID: 38085098, where they combine Bayesian variable selection methods for genetic parameter estimation, genomic prediction, and GWAS.
With HEGP anyone can freely hand over genotype and phenotype data to anyone else and have them reproduce the results. Even better, the encoding can be stacked. So the second party can encrypt and combine their data independently and give it to a third party.
So any party can use our data to add it their own analysis. And they can share their data back with us. As such, genotype data can be stored on (public) servers and GWA analysis can be reproduced.
This is a breakthrough in FAIR data sharing and contrasts greatly with current practice of hiding/protecting genotype data and only providing summary statistics. One example is an important UK Biobank depression study where genetic markers are presented with their statistical significance. We can only assume that this study can be reproduced by a group of researchers having access to the original data. The truth is that these outcomes can not be reproduced by you or me! HEGP will make data sharing and reproducible analysis a reality.
Cite
Mott R, Fischer C, Prins P, Davies RW. Private Genomes and Public SNPs: Homomorphic Encryption of Genotypes and Phenotypes for Shared Quantitative Genetics. Genetics. 2020 Jun;215(2):359-372. doi: 10.1534/genetics.120.303153. Epub 2020 Apr 23. PMID: 32327562; PMCID: PMC7268998.
Simple explanation
Homomorphic encryption is a mathematical translation of data into an
encrypted form where the result of a computation is the same for the
unencrypted and encrypted forms. With HEGP a matrix of data is
transformed by a high-dimensional random linear orthogonal
transformation key as described in the paper (open access) and
visualised in below animation (hit ). The resulting matrix
scrambles the data while preserving the 'shape of the data' for
analysis.  One way to think about this is that when a Rubik cube gets rotated the
fields change colour, but the object still maintains its shape as a
three dimensional cube. With DNA the genotype/phenotype shape is
typically used to predict associations between genotypes and
phenotypes.
One way to think about this is that when a Rubik cube gets rotated the
fields change colour, but the object still maintains its shape as a
three dimensional cube. With DNA the genotype/phenotype shape is
typically used to predict associations between genotypes and
phenotypes.  An example of a phenotype is a preference for
strawberry taste. An example of an associated genotype is a DNA
encoded olfactory receptor.
An example of a phenotype is a preference for
strawberry taste. An example of an associated genotype is a DNA
encoded olfactory receptor.
Here we display a typical example of genome-wide association (GWA) of phenotype against genotype:
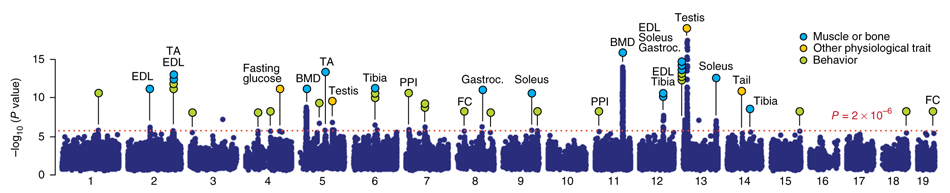
Genes (on chromosomes) involved in some trait are marked. This is the backbone computation for finding genes involved in some trait and pursued in the UK Biobank involving half a million subjects. To find associations GWA is applied to find genes involved in, for example, cancer or COVID-19 mortality; i.e., the first step towards finding causality and potentially better treatments.
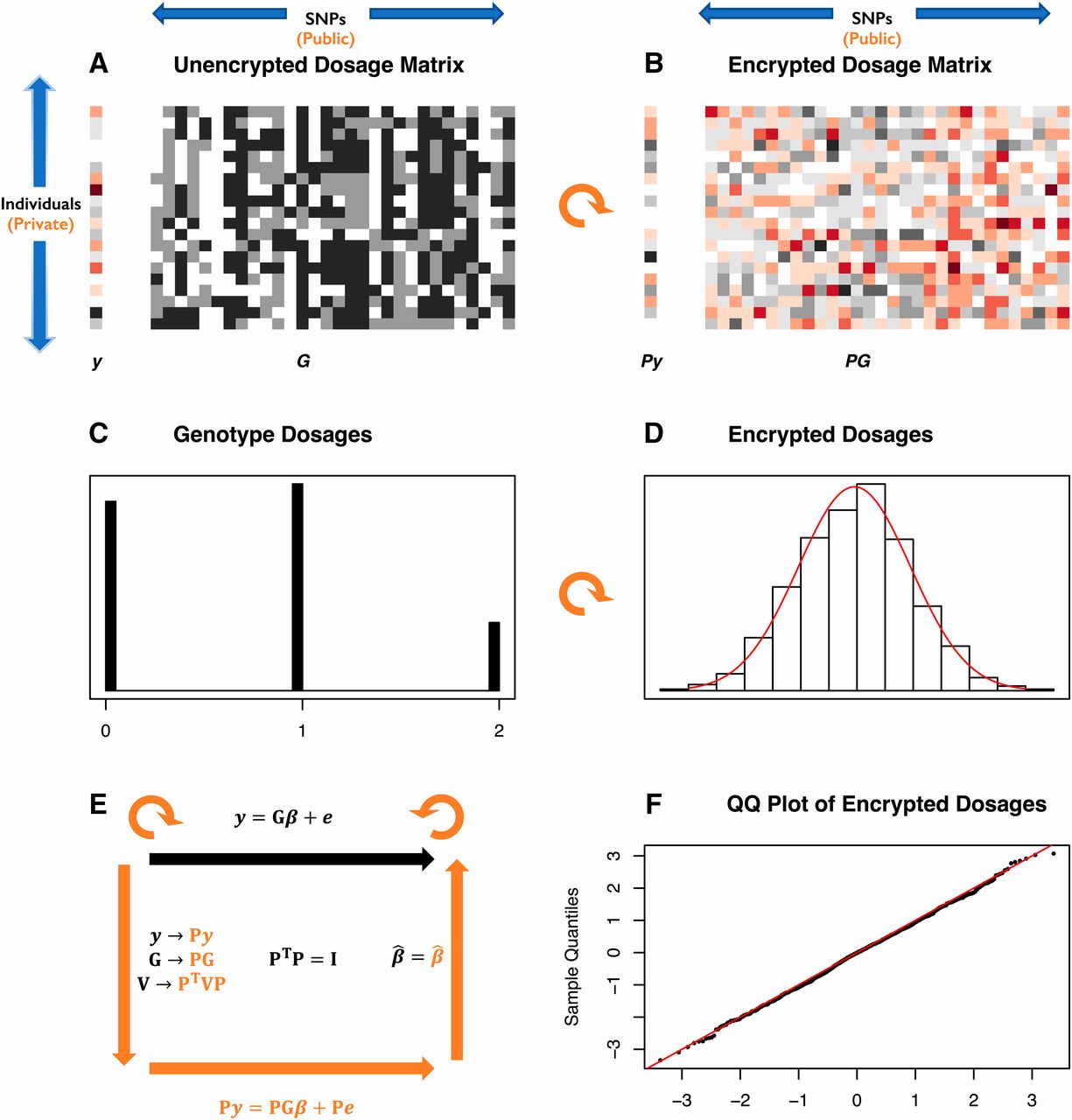
In above image data is shown before and after encryption. The unencrypted data contains three values while the encrypted data shows a normal distribution.
Enigma and why a challenge?
The Enigma machine is an encryption device developed and used in the
20th century to protect commercial, diplomatic and military
communication. It was employed extensively by Nazi Germany during
World War II, in all branches of the German military (source
wikipedia).  Enigma
encrypted text by a transformation and was cracked by the Polish
Cipher Bureau in 1932 and the crack was used by the allied forces to
win the war. To ascertain HEGP is bullet proof, unlike ENIGMA, we
invite the algorithmic inclined to crack the code and make HEGP
history (one way or the other).
Enigma
encrypted text by a transformation and was cracked by the Polish
Cipher Bureau in 1932 and the crack was used by the allied forces to
win the war. To ascertain HEGP is bullet proof, unlike ENIGMA, we
invite the algorithmic inclined to crack the code and make HEGP
history (one way or the other).